Ecosystem
Ecosystem#
What do we mean by an “ecosystem” of software projects?
Wikipedia defines an ecosystem as
a community of living organisms in conjunction with the nonliving components of their environment, interacting as a system
A few key ideas that are common among living ecosystems and an “ecosystem” of software projects are:
community
a common environment
interaction and interoperability
reliance and interdependence
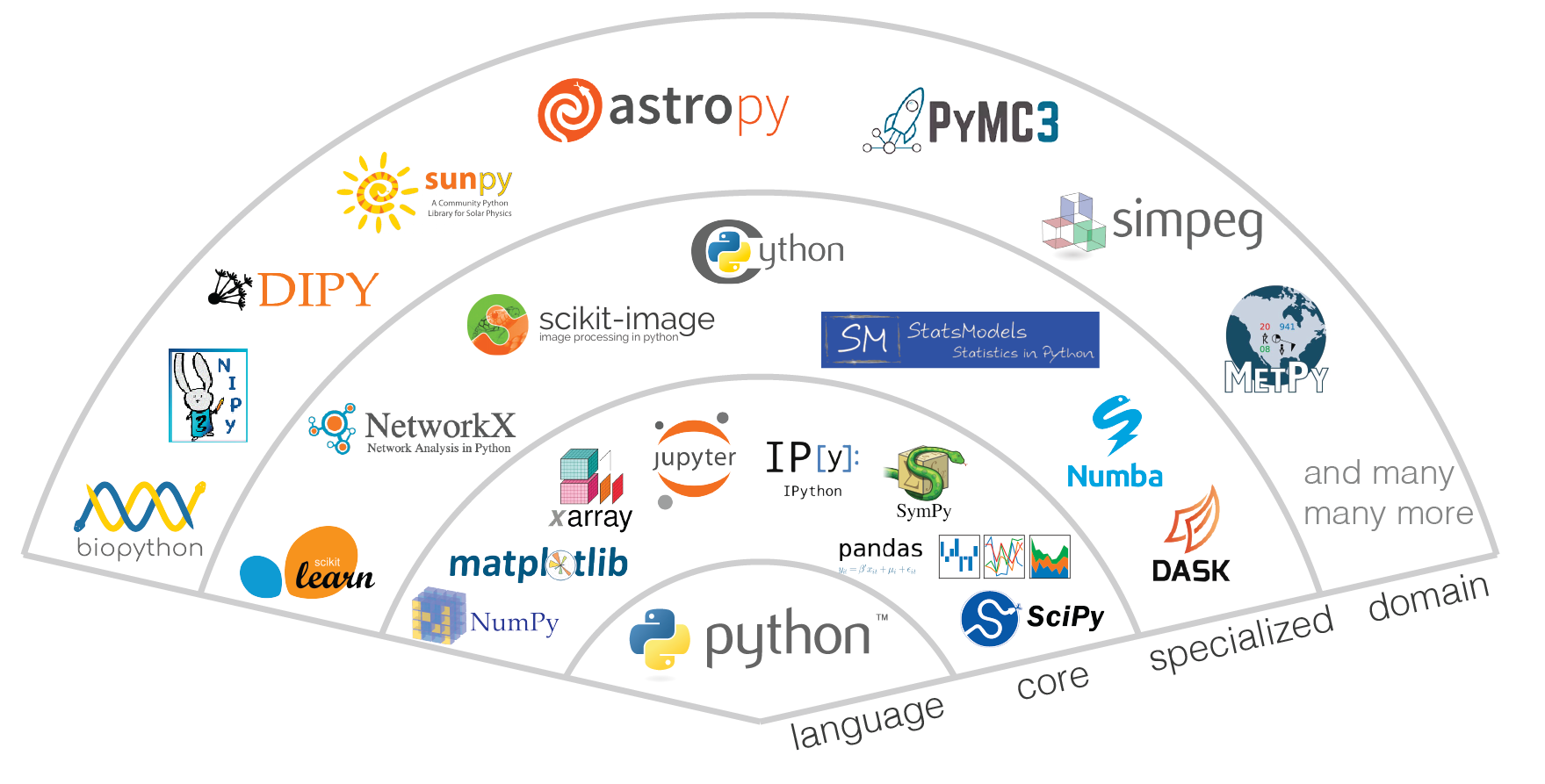
The term “ecosystem” is often used to describe the modern open-source scientific software. In biology, the term “ecosystem” is defined as a biological community of interacting organisms and their physical environment. Modern open-source scientific software development occurs in a similarly interconnected and interoperable fashion. The act of importing a software package signals a reliance of one part of the system on the functionality and role of another. Projects in this ecosystem lie somewhere along the spectrum from “foundational” projects which serve a wide community and are relied on by increasingly specialized projects, which eventually may be specialized enough to serve a specific scientific domain. Figure XX illustrates this concept of a software stack for the scientific python ecosystem. At the base of the stack is the language, in this case Python. Upon this sits the layer of “core” software packages. These packages provide functionality that is generically useful throughout the scientific ecosystem; for example Numpy for n-dimensional arrays [Harris et al., 2020], Matplotlib for plotting [Hunter, 2007], Pandas for data frames [McKinney, 2010, Pandas development team, 2020], and Jupyter for interactive computing [Perez and Granger, 2007]. The next layer consists of specialized projects such as Scikit-learn for machine learning [Pedregosa et al., 2011] or Dask for parallel computation [Dask Development Team, 2016]; these projects are relied on by others in the ecosystem to solve a general suite of computational tasks. The top layer is that of domain-specific projects, which are tailored to serve a distinct community of researchers and the questions they are pursuing. There are not necessarily hard lines between these layers, as projects mature, they may move between these layers, and some communities might organize projects differently.
- HMvdW+20
Charles R. Harris, K. Jarrod Millman, Stéfan J. van der Walt, Ralf Gommers, Pauli Virtanen, David Cournapeau, Eric Wieser, Julian Taylor, Sebastian Berg, Nathaniel J. Smith, Robert Kern, Matti Picus, Stephan Hoyer, Marten H. van Kerkwijk, Matthew Brett, Allan Haldane, Jaime Fernández del Río, Mark Wiebe, Pearu Peterson, Pierre Gérard-Marchant, Kevin Sheppard, Tyler Reddy, Warren Weckesser, Hameer Abbasi, Christoph Gohlke, and Travis E. Oliphant. Array programming with NumPy. Nature, 585(7825):357–362, sep 2020. URL: http://www.nature.com/articles/s41586-020-2649-2, doi:10.1038/s41586-020-2649-2.
- Hun07
John D. Hunter. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering, 9(3):90–95, 2007. URL: http://ieeexplore.ieee.org/document/4160265/, doi:10.1109/MCSE.2007.55.
- McK10
Wes McKinney. Data Structures for Statistical Computing in Python. In Stéfan van der Walt and Jarrod Millman, editors, Proceedings of the 9th Python in Science Conference, 56–61. 2010. doi:10.25080/Majora-92bf1922-00a.
- Pdt20
The Pandas development team. pandas-dev/pandas: Pandas. feb 2020. URL: https://doi.org/10.5281/zenodo.3509134, doi:10.5281/zenodo.3509134.
- PVG+11
F Pedregosa, G Varoquaux, A Gramfort, V Michel, B Thirion, O Grisel, M Blondel, P Prettenhofer, R Weiss, V Dubourg, J Vanderplas, A Passos, D Cournapeau, M Brucher, M Perrot, and E Duchesnay. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12:2825–2830, 2011.
- PG07
Fernando Perez and Brian E. Granger. IPython: A System for Interactive Scientific Computing. Computing in Science & Engineering, 9(3):21–29, 2007. URL: http://ieeexplore.ieee.org/document/4160251/, doi:10.1109/MCSE.2007.53.
- DaskDTeam16
Dask Development Team. Dask: Library for dynamic task scheduling. 2016. URL: https://dask.org.
